MapRRCon Analysis of ENCODE ChIP-seq Datasets on LINE-1
About
MapRRCon database contains Transcription Factor (TF) enrichment profiles on various families of LINE-1 retrotransposons.
This website is designed to visualize coverage, peaks and motifs of specific TFs on L1. We also provide TF binding profile comparisons of multiple L1 families on a subset of hESC datasets.
(more cell lines will be available soon)
Search
To start your search, please enter a name of transcription factor in the search box.
For more detailed instructions please click the help button located in the top right corner of the page.
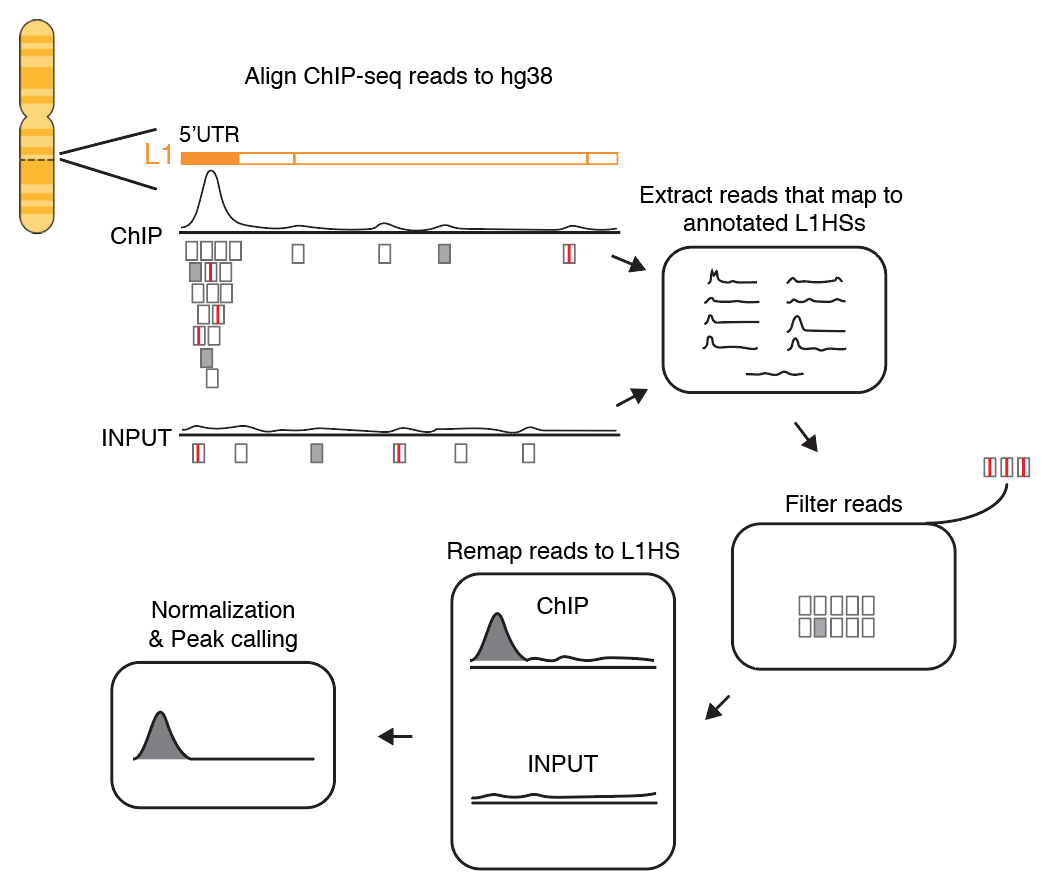
All the ChIP-seq data is available from the ENCODE website.
The methodology and analyzed data is described in the following paper:
https://www.ncbi.nlm.nih.gov/pubmed/29802231
Click here to download the scripts used by MapRRCon